Machine Learning with R
Chien-Lan Hsueh 2022-07-17
Machine Learning with R
In the last decades, there are many newly developed algorithms for machine learning. Neural networks and ensemble methods are most attractive to me. Among the tree-based ensemble learning methods we covered in this course. Random forests is the most interesting one to me.
Random forests is an ensemble method based on decision trees. It inherits many advantages from generic tree-based methods including easy and fast to implement, handling missing values, robust to outliers and automatic including interactions. Furthermore, it is based on bagging with randomly selected features in tree creation. This added randomness reduces the correlation between the sampled trees and effectively prevents highly correlated trees if there exists a strong predictor. At work, I use it more and more often to model high dimensional data.
Example
In this blog, I complete what I left off in ST558 project 2. Because of the unexpected long computing time and my sidetracking to figure out how to knit and include child documents in rmarkdown, we didn’t make enough efforts in explaining the ML methods we used. I will take this chance to do so while comparing the performance of random forests and a method not included in the project, K nearest neighbors (KNN).
First, let’s reuse the helper functions we developed in project 2.
fit_model() is a wrapper function of caret::train() with calculation
of performance metrics (depending on the data type of the response).
Some small modifications are made including making the plot option on by
default in the function call fit_mode(..., plot = TRUE) and fixing the
argument orders in caret::postResample(pred, obs,..) and
caret::confusionMatrix(data, reference,..).
Arguments:
formula: formuladf_train: training setdf_test: test setmethod: classification or regression model to usepreProcess: pre-processing of the predictorstrControl: a list of values that define how train actstuneGrid: a data frame with possible tuning valuesplot: whether to plot parameter and metric...: arguments passed to the classification or regression routineReturned Value: a performance metric (for a numeric response) or confusion matrix (for a categorical response)
plot_modelinfo() is a helper function called in fit_model() to plot
the performance metrics vs. the hyperparameter in modeling tuning
processes. This feature was turned off in the project 2.
Arguments:
fit: model fit, a list from the returned classtrainplot_wrt: which hyperparameter to plot (the column location in thetuneGrid)
# a wrapper function to train a model with train set and calculate the model performance on test set
fit_model <- function(
formula, df_train, df_test, method,
preProcess = c("center", "scale"),
trControl = trainControl(),
tuneGrid = NULL,
plot = TRUE, ... ){
# timer - start
proc_timer <- proc.time()
# train model
fit <- train(
form = formula,
data = df_train,
method = method,
preProcess = c("center", "scale"),
trControl = trControl,
tuneGrid = tuneGrid, ...)
# timer - report time used
timer <- proc.time() - proc_timer
print(timer)
# print the best tune if there is a tuning parameter
if(is.null(tuneGrid)){
print("No tuning parameter")
} else {
# print the best tune
print("The best tune is found with:")
print(glue("\t{names(fit$bestTune)} = {fit$bestTune[1,]}"))
if(plot) plot_modelinfo(fit)
}
# make prediction on test set
pred <- predict(fit, newdata = df_test)
# return performance metric or confusion matrix depending on response type
if(is.numeric(pred)){
# numeric response
performance <- postResample(pred, obs = df_test[, 1])
# print performance metrics
print("Performance metrics:")
print(performance)
# return the performance metric
return(list(method = method, performance = performance, timer = timer))
} else if(is.factor(pred)){
# categorical response
cfm <- confusionMatrix(df_test[, 1], pred)
# print confusion matrix and accuracy
print("Confusion table:")
print(cfm$table)
print(glue("Accuracy = {cfm$overall['Accuracy']}"))
# return the confusion matrix
return(list(method = method, performance = cfm, timer = timer))
}
}
# a helper function to plot the metric vs. the tuning parameter
plot_modelinfo <- function(fit, plot_wrt = 1){
# get model info
model <- fit$modelInfo$label
parameter <- fit$modelInfo$parameters$parameter
description <- fit$modelInfo$parameters$label
# plot parameter vs metrics
p <- fit$results %>%
select(-setdiff(parameter, names(fit$results)[plot_wrt])) %>%
rename_at(1, ~"x") %>%
pivot_longer(cols = -1, names_to = "Metric") %>%
ggplot(aes(x, value, color = Metric)) +
geom_point() +
geom_line() +
facet_grid(rows = vars(Metric), scales = "free_y") +
labs(
title = glue("{model}: Hyperparameter Tuning"),
x = glue("{parameter} ({description})")
)
print(p)
return(p)
}
This time, I use GermanCredit data set from caret package. Unlike
the numeric response we had in the project 2, the response Class is a
categorical response. It is used to classify the bank clients into two
groups based on their credit worthiness: good or bad. After a quick EDA
(not shown here), there are two variables containing only one single
level and thus they are removed. A 70/30 split is used to create
training and test sets.
data("GermanCredit")
df <- GermanCredit %>%
mutate(Class = factor(Class)) %>%
relocate(Class) %>%
# remove these variables since it only contains one level
select(-Personal.Female.Single, -Purpose.Vacation)
# split train/test sets
set.seed(90)
trainIndex <- createDataPartition(df$Class, p = 0.7, list = FALSE)
df_train <- df[trainIndex, ]
df_test <- df[-trainIndex, ]
Another thing I do differently here is how I save the test results. Instead of creating separate variables to save the results like we did in the project 2, a list will be used. This way I can avoid the mistake we made in the project 2 (forgot to uncomment the random forest result in the comparison section) by not hard-coded the comparison part.
# initiate a list to save the results
results <- tibble()
KNN
KKN is a supervised learning algorithm. It calculates distance between each data point. Some popular distance metrics used in KNN including Euclidean distance, Manhattan distance, Minkowski distance, cosine distance and Hamming distance. Then the distances are used to find the closet neighbors and the classification is decided by majority vote. The hyperparameter used to tune the models in cross validation is the number of neighbors (from 1 to 10 in the search grid). To get a fair comparison for all the models included in this study, I use 10-fold cross validation and repeat it 3 times to search the specified hyperparameter to get the models with the best performance metric - accuracy. Once the best model is found, that model is then used to make a prediction on the test set to test its performance (accuracy again).
The first model is K nearest neighbor (KNN) using Class::knn() with
Euclidean distance:
# configure the model to use
name <- "KNN Euclidean"
method <- "knn"
tuneGrid <- expand.grid(k = 1:15)
# fit the model
fit <- fit_model(
Class ~ ., df_train, df_test, method = method,
trControl = trainControl(method = "repeatedcv", number = 10, repeats = 3),
tuneGrid = tuneGrid
)
## user system elapsed
## 6.93 0.12 7.06
## [1] "The best tune is found with:"
## k = 14

## [1] "Confusion table:"
## Reference
## Prediction Bad Good
## Bad 18 72
## Good 12 198
## Accuracy = 0.72
# save the result
results <-
tibble(
name = name,
method = method,
accuracy = fit$performance$overall["Accuracy"],
kappa = fit$performance$overall["Kappa"],
time_elapsed = fit$timer["elapsed"]
) %>%
bind_rows(results)
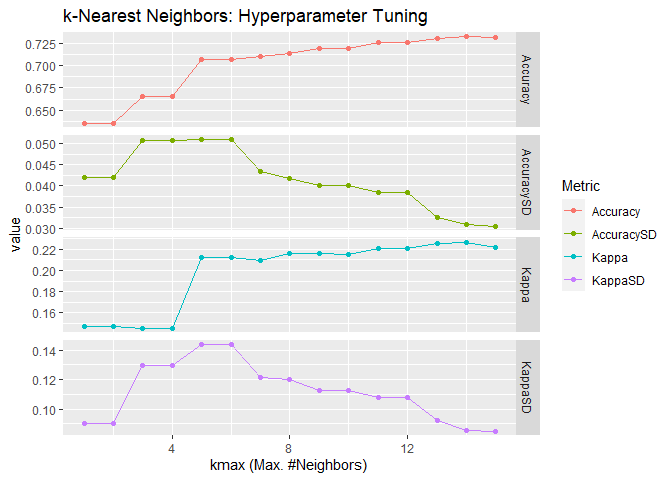
The second model is still KNN but I use kknn:kknn() with Minkowski
distance. Although it is capable to do weighted KNN, I use the standard
un-weighted KNN with kernel = "rectangular" for a fair comparison with
the previous KNN model. I also use the default value for the parameter
of Minkowski distance: distance = 2 which is Euclidean distance.
# configure the model to use
name <- "KNN Euclidean"
method <- "kknn"
tuneGrid <- expand.grid(kmax = 1:15, distance = 2, kernel = "rectangular")
# fit the model
fit <- fit_model(
Class ~ ., df_train, df_test, method = method,
trControl = trainControl(method = "repeatedcv", number = 10, repeats = 3),
tuneGrid = tuneGrid
)
## user system elapsed
## 70.23 0.22 70.53
## [1] "The best tune is found with:"
## kmax = 14
## distance = 2
## kernel = 1
## [1] "Confusion table:"
## Reference
## Prediction Bad Good
## Bad 16 74
## Good 12 198
## Accuracy = 0.713333333333333
# save the result
results <-
tibble(
name = name,
method = method,
accuracy = fit$performance$overall["Accuracy"],
kappa = fit$performance$overall["Kappa"],
time_elapsed = fit$timer["elapsed"]
) %>%
bind_rows(results)
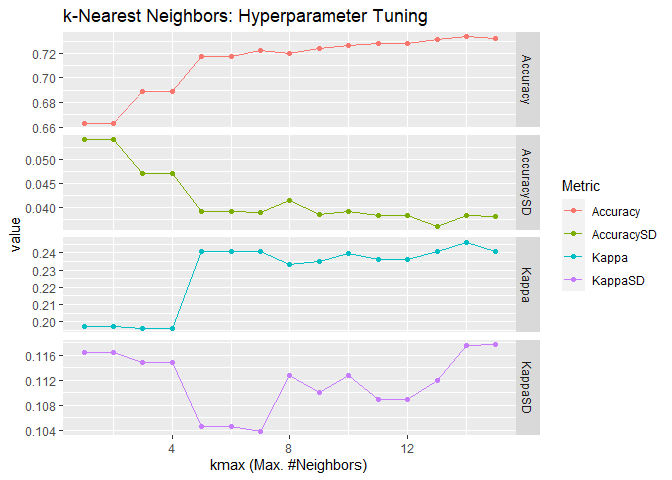
When using the parameter distance = 1, the Minkowski distance is the
Manhattan distance:
# configure the model to use
name <- "KNN Manhattan"
method <- "kknn"
tuneGrid <- expand.grid(kmax = 1:15, distance = 1, kernel = "rectangular")
# fit the model
fit <- fit_model(
Class ~ ., df_train, df_test, method = method,
trControl = trainControl(method = "repeatedcv", number = 10, repeats = 3),
tuneGrid = tuneGrid
)
## user system elapsed
## 217.93 0.21 218.23
## [1] "The best tune is found with:"
## kmax = 14
## distance = 1
## kernel = 1
## [1] "Confusion table:"
## Reference
## Prediction Bad Good
## Bad 28 62
## Good 15 195
## Accuracy = 0.743333333333333
# save the result
results <-
tibble(
name = name,
method = method,
accuracy = fit$performance$overall["Accuracy"],
kappa = fit$performance$overall["Kappa"],
time_elapsed = fit$timer["elapsed"]
) %>%
bind_rows(results)
Random Forests
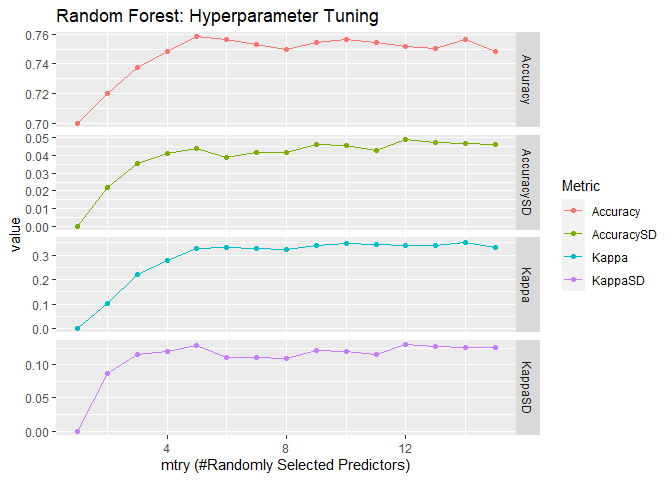
The third mode is our first random forest model and is done with
randomForest::randomForest(). I use same 10-fold cross validation with
3 repeats to search the hyperparameter mtry, number of randomly
selected predictors in each tree. The one with best accuracy will then
be used for prediction on the test set to get its performance
(accuracy).
# configure the model to use
name <- "Random Forests"
method <- "rf"
tuneGrid <- expand.grid(mtry = 1:15)
# fit the model
fit <- fit_model(
Class ~ ., df_train, df_test, method = method,
trControl = trainControl(method = "repeatedcv", number = 10, repeats = 3),
tuneGrid = tuneGrid
)
## user system elapsed
## 350.29 3.53 353.93
## [1] "The best tune is found with:"
## mtry = 5
## [1] "Confusion table:"
## Reference
## Prediction Bad Good
## Bad 30 60
## Good 10 200
## Accuracy = 0.766666666666667
# save the result
results <-
tibble(
name = name,
method = method,
accuracy = fit$performance$overall["Accuracy"],
kappa = fit$performance$overall["Kappa"],
time_elapsed = fit$timer["elapsed"]
) %>%
bind_rows(results)
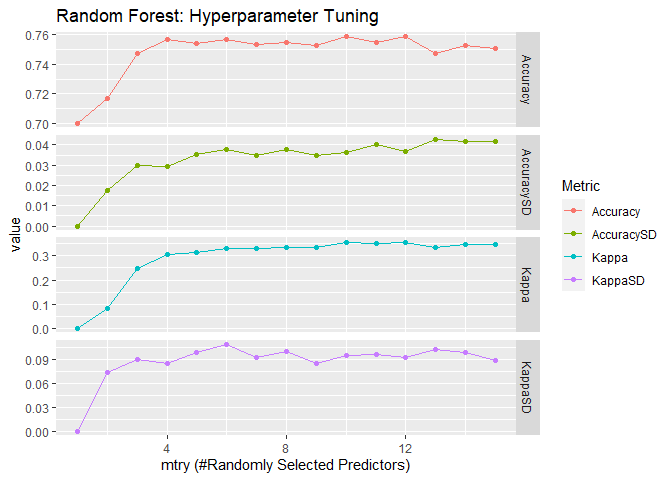
ranger::ranger() is another popular implementation of random forests.
It claims to have a faster speed and suitable for high dimensional data.
I keep tuning the same hyperparameter mtry and use gini impurity as
the splitting rule. For classification, the default value for
min.node.size is 1.
# configure the model to use
name <- "Random Forests"
method <- "ranger"
tuneGrid <- expand.grid(mtry = 1:15, splitrule = "gini", min.node.size = 1)
# fit the model
fit <- fit_model(
Class ~ ., df_train, df_test, method = method,
trControl = trainControl(method = "repeatedcv", number = 10, repeats = 3),
tuneGrid = tuneGrid
)
## user system elapsed
## 157.14 1.67 32.81
## [1] "The best tune is found with:"
## mtry = 10
## splitrule = 1
## min.node.size = 1
## [1] "Confusion table:"
## Reference
## Prediction Bad Good
## Bad 32 58
## Good 13 197
## Accuracy = 0.763333333333333
# save the result
results <-
tibble(
name = name,
method = method,
accuracy = fit$performance$overall["Accuracy"],
kappa = fit$performance$overall["Kappa"],
time_elapsed = fit$timer["elapsed"]
) %>%
bind_rows(results)
Comparison
Both KNN and random forests use voting and that makes it interesting to
see the comparison. In each learning method, we tune one hyperparameter
in the range of 1:15 using 3 repeats of 10-fold cross validation to
choose the best model with the highest accuracy. The best model is then
used to make predication on test set. We can then compare their accuracy
(together with another performance metric, Cohen’s
Kappa) and the time
used:
# sort the results
results %>%
arrange(desc(accuracy), time_elapsed)
## # A tibble: 5 × 5
## name method accuracy kappa time_elapsed
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 Random Forests rf 0.767 0.340 354.
## 2 Random Forests ranger 0.763 0.343 32.8
## 3 KNN Manhattan kknn 0.743 0.282 218.
## 4 KNN Euclidean knn 0.72 0.176 7.06
## 5 KNN Euclidean kknn 0.713 0.150 70.5
Some findings:
class:knn()is faster thankknn:kknn()ranger::ranger()is faster thanrandomForest::randomForest()as it claimed- Based on accuracy and computing time,
ranger::ranger()is the best method - and I sidetracked again and spent too much time on this blog. Need to go back to work on ST501 HW7 NOW!